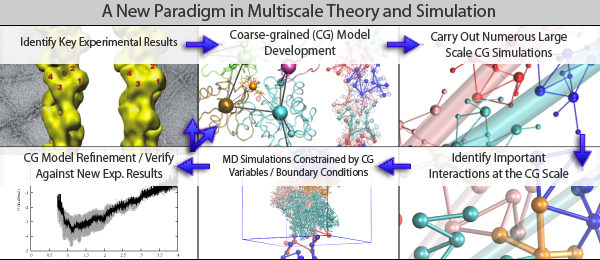
Developing and applying new theoretical and computational methods to study complex condensed phase systems
Gregory A. Voth

Haig P. Papazian Distinguished Service Professor
Department of Chemistry
Google Scholar Page
Material for Download
- OpenMSCG - Open-source software for multiscale coarse-graining modeling
- RAPTOR - Software for reactive molecular dynamics simulations
Research Highlights
Static and Dynamic Correlations in Water: Comparison of Classical Ab Initio Molecular Dynamics at Elevated Temperature with Path Integral Simulations at Ambient Temperature
It is a common practice in ab initio molecular dynamics (AIMD) simulations of water to use an elevated temperature to overcome the overstructuring and slow diffusion predicted by most current density functional theory (DFT) models. The simulation results obtained in this distinct thermodynamic state are then compared with experimental data at ambient temperature based on the rationale that a higher temperature effectively recovers nuclear quantum effects (NQEs) that are missing in the classical AIMD simulations. In this work, we systematically examine the foundation of this assumption for several DFT models as well as for the many-body MB-pol model. We find for the cases studied that a higher temperature does not correctly mimic NQEs at room temperature, which is especially manifest in significantly different three-molecule correlations as well as hydrogen bond dynamics. In many of these cases, the effects of NQEs are the opposite of the effects of carrying out the simulations at an elevated temperature.
Ion permeation, selectivity, and electronic polarization in fluoride channels
Fluoride channels (Flucs) export toxic F− from the cytoplasm. Crystallography and mutagenesis have identified several conserved residues crucial for fluoride transport, but the permeation mechanism at the molecular level has remained elusive. Herein, we have applied constant-pH molecular dynamics and free-energy-sampling methods to investigate fluoride permeation through a Fluc protein from Escherichia coli. We find that fluoride is facile to permeate in its charged form, i.e., F−, by traversing through a non-bonded network. The extraordinary F− selectivity is gained by the hydrogen-bonding capability of the central binding site and the Coulombic filter at the channel entrance. The F− permeation rate calculated using an electronically polarizable force field is significantly more accurate compared with the experimental value than that calculated using a more standard additive force field, suggesting an essential role for electronic polarization in the F−–Fluc interactions.
Computational Studies of Lipid Droplets (Feature Article)
Lipid droplets (LDs) are intracellular organelles whose primary function is energy storage. Known to emerge from the endoplasmic reticulum (ER) bilayer, LDs have a unique structure with a core consisting of neutral lipids, triacylglycerol (TG) or sterol esters (SE), surrounded by a phospholipid (PL) monolayer and decorated by proteins that come and go throughout their complex lifecycle. In this Feature Article, we review recent developments in computational studies of LDs, a rapidly growing area of research. We highlight how molecular dynamics (MD) simulations have provided valuable molecular-level insight into LD targeting and LD biogenesis. Additionally, we review the physical properties of TG from different force fields compared with experimental data. Possible future directions and challenges are discussed.
Strain and rupture of HIV-1 capsids during uncoating
The mature capsids of HIV-1 are transiently stable complexes that self-assemble around the viral genome during maturation, and uncoat to release preintegration complexes that archive a double-stranded DNA copy of the virus in the host cell genome. However, a detailed view of how HIV cores rupture remains lacking. Here, we elucidate the physical properties involved in capsid rupture using a combination of large-scale all-atom molecular dynamics simulations and cryo-electron tomography. We find that intrinsic strain on the capsid forms highly correlated patterns along the capsid surface, along which cracks propagate. Capsid rigidity also increases with high strain. Our findings provide fundamental insight into viral capsid uncoating.
Cooperative multivalent receptor binding promotes exposure of the SARS-CoV-2 fusion machinery core
The molecular events that permit the spike glycoprotein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to bind and enter cells are important to understand for both fundamental and therapeutic reasons. Spike proteins consist of S1 and S2 domains, which recognize angiotensin-converting enzyme 2 (ACE2) receptors and contain the viral fusion machinery, respectively. Ostensibly, the binding of spike trimers to ACE2 receptors promotes dissociation of the S1 domains and exposure of the fusion machinery, although the molecular details of this process have yet to be observed. We report the development of bottom-up coarse-grained (CG) models consistent with cryo-electron tomography data, and the use of CG molecular dynamics simulations to investigate viral binding and S2 core exposure. We show that spike trimers cooperatively bind to multiple ACE2 dimers at virion-cell interfaces in a manner distinct from binding between soluble proteins, which processively induces S1 dissociation. We also simulate possible variant behavior using perturbed CG models, and find that ACE2-induced S1 dissociation is primarily sensitive to conformational state populations and the extent of S1/S2 cleavage, rather than ACE2 binding affinity. These simulations reveal an important concerted interaction between spike trimers and ACE2 dimers that primes the virus for membrane fusion and entry.
Key Factors Governing Initial Stages of Lipid Droplet Formation
Lipid droplets (LDs) are neutral lipid storage organelles surrounded by a phospholipid (PL) monolayer. LD biogenesis from the endoplasmic reticulum is driven by phase separation of neutral lipids, overcoming surface tension and membrane deformation. However, the core biophysics of the initial steps of LD formation remains relatively poorly understood. Here, we use a tunable, phenomenological coarse–grained model to study triacylglycerol (TG) nucleation in a bilayer membrane. We show that PL rigidity has a strong influence on TG lensing and membrane remodeling: when membrane rigidity increases, TG clusters remain more planar with high anisotropy but a minor degree of phase nucleation. This finding is confirmed by advanced sampling simulations that calculate nucleation free energy as a function of the degree of nucleation and anisotropy. We also show that asymmetric tension, controlled by the number of PL molecules on each membrane leaflet, determines the budding direction. A TG lens buds in the direction of the monolayer containing excess PL molecules to allow for better PL coverage of TG, consistent with the reported experiments. Finally, two governing mechanisms of the LD growth, Ostwald ripening and merging, are observed. Taken together, this study characterizes the interplay between two thermodynamic quantities during the initial LD phases, the TG bulk free energy and membrane remodeling energy.
Using Machine Learning to Greatly Accelerate Path Integral Ab Initio Molecular Dynamics
Ab Initio molecular dynamics (AIMD) has become one of the most popular and robust approaches for modeling complicated chemical, liquid, and material systems. However, the formidable computational cost often limits its widespread application in simulations of the largest-scale systems. The situation becomes even more severe in cases where the hydrogen nuclei may be better described as quantized particles using a path integral representation. Here, we present a computational approach that combines machine learning with recent advances in path integral contraction schemes, and we achieve a 2 orders of magnitude acceleration over direct path integral AIMD simulation while at the same time maintaining its accuracy.
Multiscale Simulation of an Influenza A M2 Channel Mutant Reveals Key Features of Its Markedly Different Proton Transport Behavior
The influenza A M2 channel, a prototype for viroporins, is an acid-activated viroporin that conducts protons across the viral membrane, a critical step in the viral life cycle. Four central His37 residues control channel activation by binding subsequent protons from the viral exterior, which opens the Trp41 gate and allows proton flux to the interior. Asp44 is essential for maintaining the Trp41 gate in a closed state at high pH, resulting in asymmetric conduction. The prevalent D44N mutant disrupts this gate and opens the C-terminal end of the channel, resulting in increased conduction and a loss of this asymmetric conduction. Here, we use extensive Multiscale Reactive Molecular Dynamics (MS-RMD) and quantum mechanics/molecular mechanics (QM/MM) molecular dynamics simulations with an explicit, reactive excess proton to calculate the free energy of proton transport in this M2 mutant and to study the dynamic molecular-level behavior of D44N M2. We find that this mutation significantly lowers the barrier of His37 deprotonation in the activated state and shifts the barrier for entry to the Val27 tetrad. These free energy changes are reflected in structural shifts. Additionally, we show that the increased hydration around the His37 tetrad diminishes the effect of the His37 charge on the channel’s water structure, facilitating proton transport and enabling activation from the viral interior. Altogether, this work provides key insight into the fundamental characteristics of PT in WT M2 and how the D44N mutation alters this PT mechanism, and it expands understanding of the role of emergent mutations in viroporins.
Molecular interactions of the M and E integral membrane proteins of SARS-CoV-2
Specific lipid–protein interactions are key for cellular processes, and even more so for the replication of pathogens. The COVID-19 pandemic has drastically changed our lives and caused the death of nearly four million people worldwide, as of this writing. SARS-CoV-2 is the virus that causes the disease and has been at the center of scientific research over the past year. Most of the research on the virus is focused on key players during its initial attack and entry into the cellular host; namely the S protein, its glycan shield, and its interactions with the ACE2 receptors of human cells. As cases continue to rise around the globe, and new mutants are identified, there is an urgent need to understand the mechanisms of this virus during different stages of its life cycle. Here, we consider two integral membrane proteins of SARS-CoV-2 known to be important for viral assembly and infectivity. We have used microsecond-long all-atom molecular dynamics to examine the lipid–protein and protein–protein interactions of the membrane (M) and envelope (E) structural proteins of SARS-CoV-2 in a complex membrane model. We contrast the two proposed protein complexes for each of these proteins, and quantify their effect on their local lipid environment. This ongoing work also aims to provide molecular-level understanding of the mechanisms of action of this virus to possibly aid in the design of novel treatments.
A Quantitative Paradigm for Water Assisted Proton Transport Through Proteins and Other Confined Spaces
Water-assisted proton transport through confined spaces influences many phenomena in biomolecular and nanomaterial systems. In such cases, the water molecules that fluctuate in the confined pathways provide the environment and the medium for the hydrated excess proton migration via Grotthuss shuttling. However, a definitive collective variable (CV) that accurately couples the hydration and the connectivity of the proton wire with the proton translocation has remained elusive. To address this important challenge—and thus to define a quantitative paradigm for facile proton transport in confined spaces—a CV is derived in this work from graph theory, which is verified to accurately describe water wire formation and breakage coupled to the proton translocation in carbon nanotubes and the Cl−/H+ antiporter protein, ClC-ec1. Significant alterations in the conformations and thermodynamics of water wires are uncovered after introducing an excess proton into them. Large barriers in the proton translocation free-energy profiles are found when water wires are defined to be disconnected according to the new CV, even though the pertinent confined space is still reasonably well hydrated and—by the simple measure of the mere existence of a water structure—the proton transport would have been predicted to be facile via that oversimplified measure. In this paradigm, however, the simple presence of water is not sufficient for inferring proton translocation, since an excess proton itself is able to drive hydration, and additionally, the water molecules themselves.
Preservation of HIV-1 Gag Helical Bundle Symmetry by Bevirimat Is Central to Maturation Inhibition
The assembly and maturation of human immunodeficiency virus type 1 (HIV-1) require proteolytic cleavage of the Gag polyprotein. The rate-limiting step resides at the junction between the capsid protein CA and spacer peptide 1, which assembles as a six-helix bundle (6HB). Bevirimat (BVM), the first-in-class maturation inhibitor drug, targets the 6HB and impedes proteolytic cleavage, yet the molecular mechanisms of its activity, and relatedly, the escape mechanisms of mutant viruses, remain unclear. Here, we employed extensive molecular dynamics (MD) simulations and free energy calculations to quantitatively investigate molecular structure–activity relationships, comparing wild-type and mutant viruses in the presence and absence of BVM and inositol hexakisphosphate (IP6), an assembly cofactor. Our analysis shows that the efficacy of BVM is directly correlated with preservation of 6-fold symmetry in the 6HB, which exists as an ensemble of structural states. We identified two primary escape mechanisms, and both lead to loss of symmetry, thereby facilitating helix uncoiling to aid access of protease. Our findings also highlight specific interactions that can be targeted for improved inhibitor activity and support the use of MD simulations for future inhibitor design.
Resolving the Structural Debate for the Hydrated Excess Proton in Water
It has long been proposed that the hydrated excess proton in water (aka the solvated “hydronium” cation) likely has two limiting forms, that of the Eigen cation (H9O4+) and that of the Zundel cation (H5O2+). There has been debate over which of these two is the more dominant species and/or whether intermediate (or “distorted”) structures between these two limits are the more realistic representation. Spectroscopy experiments have recently provided further results regarding the excess proton. These experiments show that the hydrated proton has an anisotropy reorientation time scale on the order of 1–2 ps. This time scale has been suggested to possibly contradict the picture of the more rapid “special pair dance” phenomenon for the hydrated excess proton, which is a signature of a distorted Eigen cation. The special pair dance was predicted from prior computational studies in which the hydrated central core hydronium structure continually switches (O–H···O)* special pair hydrogen-bond partners with the closest three water molecules, yielding on average a distorted Eigen cation with three equivalent and dynamically exchanging distortions. Through state-of-art simulations it is shown here that anisotropy reorientation time scales of the same magnitude are obtained that also include structural reorientations associated with the special pair dance, leading to a reinterpretation of the experimental results. These results and additional analyses point to a distorted and dynamic Eigen cation as the most prevalent hydrated proton species in aqueous acid solutions of dilute to moderate concentration, as opposed to a stabilized or a distorted (but not “dancing”) Zundel cation.
Integrin-Based Mechanosensing through Conformational Activation
Cells can detect and react to the biophysical properties of the extracellular environment through integrin-based adhesion sites and adapt to the extracellular milieu in a process called mechanotransduction. At these adhesion sites, integrins connect the extracellular matrix (ECM) with the F-actin cytoskeleton and transduce mechanical forces generated by the actin retrograde flow and myosin II to the ECM through mechanosensitive focal adhesion proteins that are collectively termed the “molecular clutch.” The transmission of forces across integrin-based adhesions establishes a mechanical reciprocity between the viscoelasticity of the ECM and the cellular tension. During mechanotransduction, force allosterically alters the functions of mechanosensitive proteins within adhesions to elicit biochemical signals that regulate both rapid responses in cellular mechanics and long-term changes in gene expression. Integrin-mediated mechanotransduction plays important roles in development and tissue homeostasis, and its dysregulation is often associated with diseases.